Image: Double-strand break repair pathway models
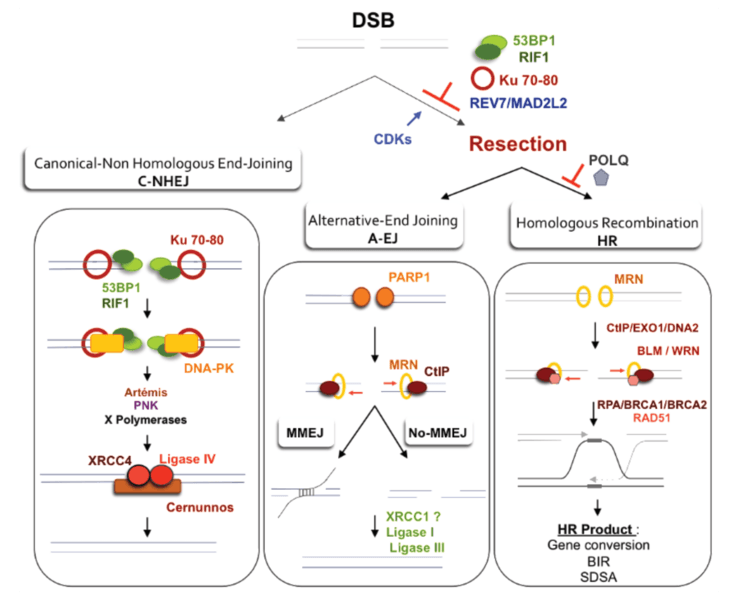
Description: (Left panel): Canonical C-NHEJ. The heterodimer Ku80-Ku70 binds to DNA ends, which then recruits DNA-PKcs. In subsequent steps, several proteins including Artemis, polynucleotide kinase (PNK), and members of the polymerase X family process the DNA ends. In the last step, ligase IV associated with its co-factors Xrcc4 and Cernunos/XLF joins the ends. (Right Panel): Resection as a common initiation step for HR and A-EJ at DSB. 53BP1, RIF1 and Ku70-80 heterodimer protect DSB ends from resection and HR and A-EJ actions. The CDK1/2-dependent phosphorylation of CtIP and EXO1 favors the initiation of resection and extension, respectively. Recently, REV7/MAD2L2 was described as an inhibitor of resection and HR, although its role in A-EJ inhibition was not directly studied and remains hypothetical. A short ssDNA resection allows for A-EJ but not homologous recombination, while a long ssDNA resection allows for both A-EJ and HR; however, HR requires the presence of homologous sequences. Recently, POLQ polymerase was shown to inhibit HR and to promote A-EJ at DSBs. A-EJ results in repair that is error-prone and is associated with deletions at the repair junctions with frequent use of microhomologies that are distant from the DSB. Alternative-EJ: Parp1 plays a role in the initiation process, and it has been proposed that a single-strand DNA resection reveals complementary microhomologies (two to four nucleotides or more in length) that can anneal, with gap-filling completing the end-joining. A-EJ is always associated with deletions at the junctions and can involve microhomologies (MMEJ or microhomologies-mediated EJ) that are distant from the DSB. Subsequently, Xrcc1 and ligase III (which can be substituted by ligase I) complete the A-EJ process. Homologous recombination: The first step, which is the initiation of resection, involves the removal of ~50–100 bases of DNA from the 5' end by the MRN complex (Mre11-Rad50-Nbs1) in conjunction with CtIP. The second step, resection extension, is carried out by two alternate pathways involving either the 5' to 3' exonuclease EXO1 or the helicase-topoisomerase complex BLM-TOPIIIα-RMI1-2 in concert with the nuclease CtIP/DNA2. WRN helicase has also been shown to act with CtIP and to stimulate resection in human cells.[1]
Title: Double-strand break repair pathway models
Credit: https://doi.org/10.3390/genes6020267
Author: Camille Gelot, Indiana Magdalou, and Bernard S. Lopez
Usage Terms: Creative Commons Attribution 4.0
License: CC BY 4.0
License Link: https://creativecommons.org/licenses/by/4.0
Attribution Required?: Yes
Image usage
The following page links to this image:

