ENCODE facts for kids
 |
|
| Content | |
|---|---|
| Description | Whole-genome database |
| Contact | |
| Research center | Stanford University |
| Laboratory | Stanford Genome Technology Center: Cherry Lab; Formerly: University of California, Santa Cruz |
| Authors | Eurie L. Hong and 17 others |
| Primary citation | PubMed |
| Release date | 2010 |
The Encyclopedia of DNA Elements (ENCODE) is a big science project. Its main goal is to create a complete map of all the important parts of the human genome. Think of it like building a detailed instruction manual for our DNA.
ENCODE also helps other scientists. It creates a huge collection of information about our genes. This includes data, computer programs, and tools for studying DNA. All of this helps researchers learn more about our bodies.
The project is always growing. It adds more types of cells and more kinds of information. It even includes studies on mouse DNA now.
Contents
What is ENCODE?
ENCODE stands for "Encyclopedia of DNA Elements." It is a large public research project. The project aims to find all the "functional elements" in the human genome. Functional elements are the parts of our DNA that do something important.
This project helps other scientists. It gives them lots of data and tools. This helps them understand how our genes work. It also helps them learn about diseases.
The project started in 2003. It is a follow-up to the Human Genome Project. That project mapped all our genes. ENCODE wants to find out what all those genes actually *do*.
Why is ENCODE Important?
Humans have about 20,000 genes that make proteins. These genes make up only about 1.5% of our DNA. For a long time, scientists called the rest of our DNA "junk." They thought it did not do anything important.
The ENCODE project wants to find out what the other 98.5% of our DNA does. Many parts of this "non-coding" DNA control how our genes work. These parts can turn genes on or off. They can also change how much protein a gene makes.
Changes in these control parts can cause diseases. By finding these control elements, ENCODE helps us understand diseases better. It can also help scientists find new ways to treat them.
Who Works on ENCODE?
The ENCODE project involves many scientists. They come from research groups all over the world. Most of them get money from the US National Human Genome Research Institute (NHGRI).
The project started small. It had only a few research groups. But it grew quickly. Now, hundreds of scientists from many labs work together. They share all their findings with everyone. This helps other researchers use the information freely.
How ENCODE Works: The Project Phases
The ENCODE project has happened in different stages, called phases. Each phase had specific goals.
Phase 1: The Pilot Project
The first phase was like a test run. It started in 2003. Scientists wanted to find the best ways to study DNA. They tested different methods to find important parts of the human genome.
They looked at about 1% of the human genome. This was about 30 million DNA letters. All the data they found was quickly shared with other scientists.
What They Found in Phase 1
The pilot phase was a success. Scientists learned many new things about our DNA. Here are some key discoveries:
- Most of our DNA is active. It makes at least one type of RNA.
- They found many new types of RNA that do not make proteins.
- They found many new "start sites" for genes. These are like the "on" switches for genes.
- They learned that how DNA is packaged in a cell affects when genes are active.
- About 5% of our DNA has stayed the same over millions of years. This means it is very important.
- Many parts of our DNA are active but do not seem to be important for survival. These might be like a "warehouse" of useful parts for future changes.
Phase 2: The Production Phase
After the pilot project, ENCODE started its "production phase" in 2007. The goal was to study the *entire* human genome. This was a much bigger job.
Many more scientists joined the project. They used new, faster machines to read DNA. They created a huge amount of data. By 2010, they had over 1,000 sets of data. This data showed which parts of DNA make RNA. It also showed which parts control genes.
What They Found in Phase 2
In 2012, ENCODE released many new findings. They published 30 papers at once! Here are some of the most important discoveries:
- Over 80% of the human genome is active. This means it does something important. This was a big surprise!
- Most of our DNA is very close to a control point. This means almost all of our DNA is involved in some activity.
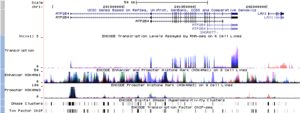
- They found almost 400,000 regions that act like "enhancers." These are like volume controls for genes. They also found over 70,000 "promoters," which are like "on" switches.
- They found that changes in our DNA that do not make proteins can still cause diseases. Many disease-related changes are in these non-coding parts of DNA.
The biggest finding was that much more of our DNA is active than scientists thought. This "active" DNA helps control the genes that make proteins.
Data Management
Managing all this data is a huge task. The ENCODE Data Coordination Center (DCC) collects and organizes all the information. They make sure the data is high quality. They also make sure it is easy for other scientists to use.
All ENCODE data is free and open to everyone. This helps scientists around the world use the information to make new discoveries.
Other Projects Like ENCODE
The ENCODE project has inspired other similar projects.
modENCODE Project
The modENCODE project is like ENCODE but for "model organisms." These are animals like fruit flies (Drosophila melanogaster) and tiny worms (Caenorhabditis elegans). Scientists study these animals because they are simpler than humans. It is easier to test ideas about DNA in them. This project finished its work in 2012.
modERN Project
modERN is a project that grew from modENCODE. It focuses on finding more control sites in fruit flies and worms. It started around 2015.
Genomics of Gene Regulation
This project started in 2015. It studies how gene networks work in different parts of the body. The ENCODE data center also helps share data for this project.
Roadmap Epigenomics Mapping Consortium
This project started in 2008. It aimed to create a public map of "epigenomes." The epigenome is like a layer of instructions on top of our DNA. It tells genes when and where to turn on or off. This project released its main findings in 2015. Some of its data is also available through ENCODE.
fruitENCODE Project
The fruitENCODE project is a plant version of ENCODE. It studies DNA in fleshy fruits. It looks at how DNA changes as fruit ripens.
FactorBook
FactorBook is a website that helps scientists understand ENCODE data. It is like a wiki where scientists can find information about "transcription factors." These are proteins that stick to DNA and control genes. FactorBook helps show where these proteins bind and what they do.
See also
 In Spanish: ENCODE para niños
In Spanish: ENCODE para niños
- GENCODE
- Functional genomics
- Human Genome Project
- 1000 Genomes Project
 | Roy Wilkins |
 | John Lewis |
 | Linda Carol Brown |